

As of December 2015, the total number of NGS based studies have doubled in the eye field within two years. Profiling of genomic variations is more employed than expression and genome binding profiling in vision research studies. We believe the number of scientific reports based on NGS technologies will continue to increase as NGS becomes more available and affordable. Cumulative number of biomedical research papers based on NGS technologies from 2008 to 2015 in PubMed database. The first NGS report was published a decade after the launch of human genome project. From the discovery of DNA molecule until today, substantial scientific and technical advancements in human genetics and eye field are presented in a chronological order. NGS based applications have been utilized widely in vision and other biomedical research. Since I’m still very new to this field, I would like to hear your advice.Timeline of human genetics and genomic technologies. In my next article, I will walk you through the details of pairwise sequence alignment and a few common algorithms that are being used in the field. Hope you got a basic idea about sequence data analysis.

#Technology sequence analysis software#
Furthermore, you can find a list of sequence alignment software from here.
You can find a list of software tools used for DNA sequencing from here.
#Technology sequence analysis manual#
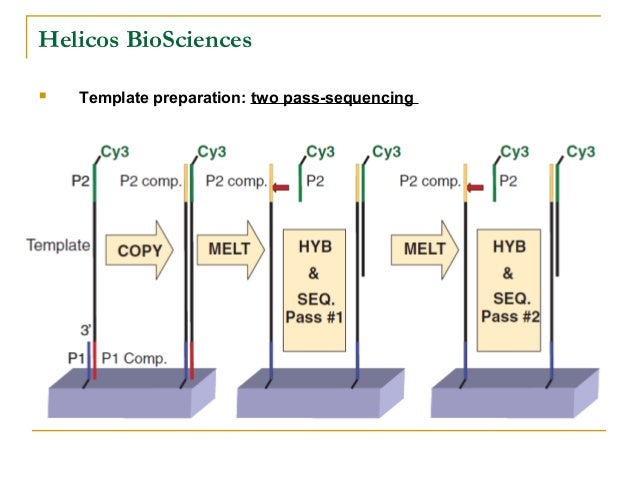
Bioinformatics has made the task of analysis much easier for biologists, by providing different software solutions and saving all the tedious manual work. Sequence data analysis has become a very important aspect in the field of genomics. These recent technologies allow us to sequence DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing, and have revolutionized the study of genomics. Next-generation sequencing (NGS), also known as high-throughput sequencing, is the collective term used to describe many different modern sequencing technologies such as, If we compare more than two sequences, it is known as multiple sequence alignment. If we compare two sequences, it is known as pairwise sequence alignment. The similarity being identified, may be a result of functional, structural, or evolutionary relationships between the sequences. Sequence alignment is a method of arranging sequences of DNA, RNA, or protein to identify regions of similarity. However, there are greedy algorithms to solve the sequence assembly problem, where experiments have proven to perform fairly well in practice.Ī common method used to solve the sequence assembly problem and perform sequence data analysis is sequence alignment. No polynomial time algorithm has yet been discovered for any NP complete problem, nor has anybody yet been able to prove that no polynomial-time algorithm exists for any of them. NP complete problems are problems whose status is unknown. It sounds very confusing and quite impossible to be carried out. Parts from another book may have also been added in, and some shreds may be completely unrecognizable. The original copy may have many repeated paragraphs, and some shreds may be modified during shredding to have typos. Furthermore, there are some extra practical issues as well. It is obvious that this task is pretty difficult. The sequence assembly problem can be compared to a real life scenario as follows.Īssume that you take many copies of a book, pass each of them through a shredder with a different cutter, and then you try to make the text of the book back together just by gluing together the shredded pieces. This problem is further complicated due to the existence of repetitive sequences in the genome as well as substitutions or mutations withing them. Given a set of sequences, find the minimal length string containing all members of the set as substrings. The sequence assembly problem can be described as follows. In order to obtain the whole genome sequence, we may need to generate more and more random reads, until the contigs match to the target genome. The process of aligning and merging fragments from a longer DNA sequence, in order to reconstruct the original sequence is known as Sequence Assembly. These contigs can be the whole target genome itself, or parts of the genome (as shown in the above figure). Special software are used to assemble these reads according to how they overlap, in order to generate continuous strings called contigs. It reads small pieces of between 0 bases, depending on the technology used. Current DNA sequencing technologies cannot read one whole genome at once. Special machines, known as sequencing machines are used to extract short random DNA sequences from a particular genome we wish to determine ( target genome).


 0 kommentar(er)
0 kommentar(er)
